YO3-3PEG-Biotin Fluorophore
| Cat. No. | G7957 | ||
| Name | YO3-3PEG-Biotin Fluorophore | ||
| Unit | 250 µM (100 µl) | ||
| Category | RNA Tracking (RNA Mango) | ||
| Description |
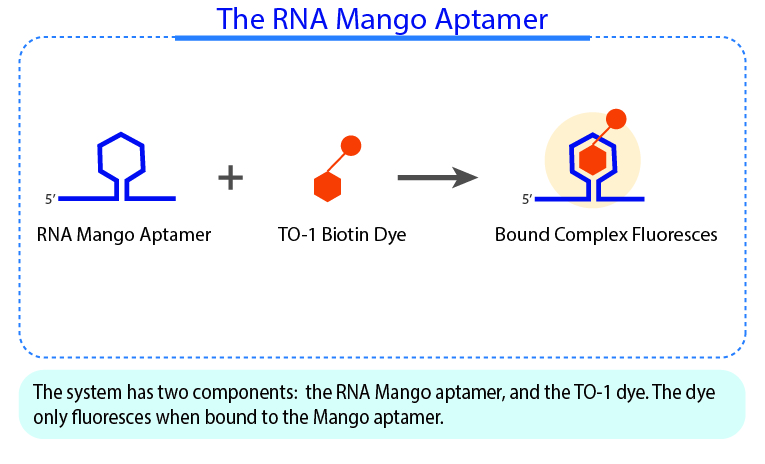
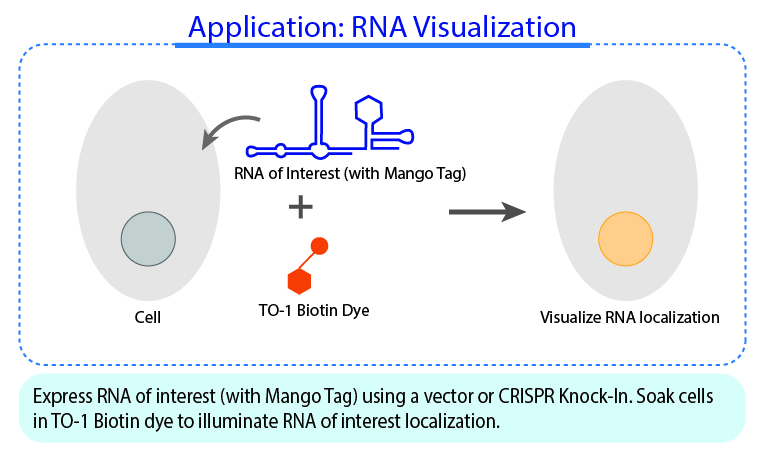
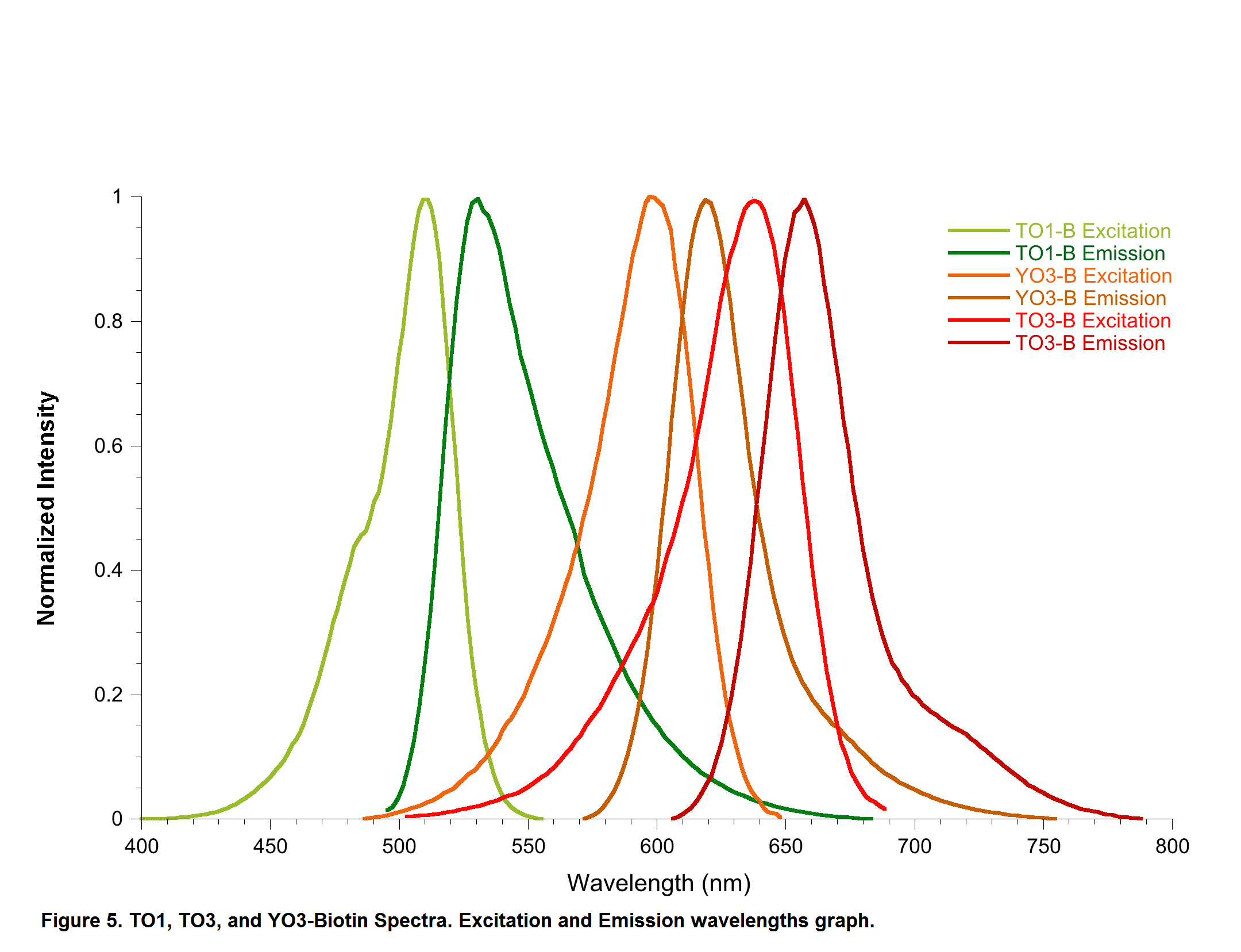
YO3-3PEG-Biotin is a small bifunctional fluorophore that has low unbound fluorescence. When bound to Mango aptamers, it exhibits peak excitation maxima of 580 nm (with additional excitation at 260 nm) and peak fluorescence emission of 620 nm. Mango aptamers enhance the fluorescence of YO3-3PEG-Biotin (binding requires KCl, 61-fold brighter with Mango III A10U), emitting in the orange region of the visible spectrum (unpublished data from Unrau Laboratory). YO3-3PEG-Biotin may serve as a FRET acceptor, when paired with GFP-emitting fluorophores1. RNA Mango technology is based on the specific binding of the RNA Mango Aptamer and a Thizole Orange (TO) bi-functional dye. The TO dye has a number of other desirable properties including:
TO3-3PEG-Biotin has binding affinities to Mango I, Mango II, Mango III, and Mango IV aptamers. Learn more about the RNA Mango technology here.
|
||
| Storage Condition | Stored at -20°C. Protect from light. | ||
| Depositor | Unrau Lab | ||
| Material Citation | If use of this material results in a scientific publication, please cite the material in the following manner: Applied Biological Materials Inc, Cat. No. G7957 |
| Can I use water with my fluorophore dyes? | |
|
We strongly recommend to use DMF, DMSO, 10% Acetonitrile, or MeOH-CH₂ Cl₂ for stability. Do no store in water as it may break down the dye.
|
| Which aptamers should I use for each RNA Mango fluorophore? | |
|
Use our RNA Mango Aptamer Systems chart to help you select the appropriate aptamer and fluorophore combination for your research needs.
|
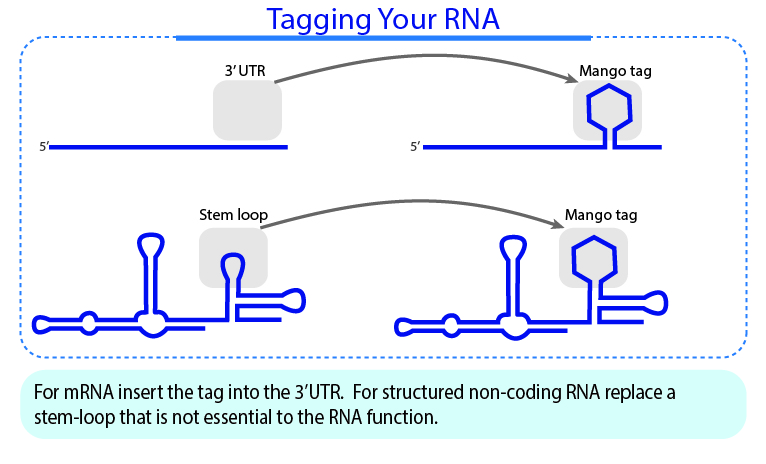
| Any tips for using RNA Mango aptamers in cellular transcripts? | |
|
We have an aptamer insertion guideline for cellular transcript, which can be found here.
|
| What is the difference between TO1-3PEG-Biotin (G955) and TO1-3PEG-Desthiobiotin (G956)? | |
|
TO1-3PEG-Biotin (G955) and TO1-3PEG-Desthiobiotin (G956) are quite similar in terms of binding and fluorescence. They are distinct in how they interact with streptavidin magnetic beads. Specifically, desthiobiotin can be displaced from them by the addition of free biotin while TO1-3PEG-Biotin cannot.
|
- Autour, A., C Y Jeng, S., D Cawte, A., Abdolahzadeh, A., Galli, A., Panchapakesan, S. S. S., Rueda, D., Ryckelynck, M., & Unrau, P. J. (2018). Fluorogenic RNA Mango aptamers for imaging small non-coding RNAs in mammalian cells. Nature communications, 9(1), 656. https://doi.org/10.1038/s41467-018-02993-8
- Trachman, R. J., & Ferré-D'Amaré, A. R. (2019). Tracking RNA with light: selection, structure, and design of fluorescence turn-on RNA aptamers. Quarterly reviews of biophysics, 52, e8. https://doi.org/10.1017/S0033583519000064
- Trachman, R. J., 3rd, Autour, A., Jeng, S. C. Y., Abdolahzadeh, A., Andreoni, A., Cojocaru, R., Garipov, R., Dolgosheina, E. V., Knutson, J. R., Ryckelynck, M., Unrau, P. J., & Ferré-D'Amaré, A. R. (2019). Structure and functional reselection of the Mango-III fluorogenic RNA aptamer. Nature chemical biology, 15(5), 472–479. https://doi.org/10.1038/s41589-019-0267-9
- Kong, K. Y. S., Jeng, S. C. Y., Rayyan, B., & Unrau, P. J. (2021). RNA Peach and Mango: Orthogonal two-color fluorogenic aptamers distinguish nearly identical ligands. RNA (New York, N.Y.), 27(5), 604–615. Advance online publication. https://doi.org/10.1261/rna.078493.120
- Cawte, A. D., Unrau, P. J., & Rueda, D. S. (2020). Live cell imaging of single RNA molecules with fluorogenic Mango II arrays. Nature communications, 11(1), 1283. https://doi.org/10.1038/s41467-020-14932-7
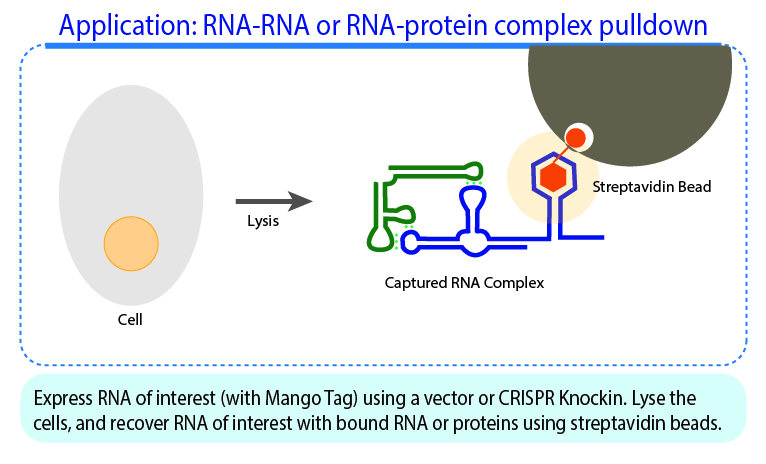
- Panchapakesan, S. S. S., Ferguson, M. L., Hayden, E. J., Chen, X., Hoskins, A. A., & Unrau, P. J. (2017). Ribonucleoprotein purification and characterization using RNA Mango. RNA (New York, N.Y.), 23(10), 1592–1599. https://doi.org/10.1261/rna.062166.117
- Mitra, J., & Ha, T. (2019). Nanomechanics and co-transcriptional folding of Spinach and Mango. Nature communications, 10(1), 4318. https://doi.org/10.1038/s41467-019-12299-y
- Shi, J., Gao, X., Tian, T., Yu, Z., Gao, B., Wen, A., You, L., Chang, S., Zhang, X., Zhang, Y., & Feng, Y. (2019). Structural basis of Q-dependent transcription antitermination. Nature communications, 10(1), 2925. https://doi.org/10.1038/s41467-019-10958-8
- Fang, C., Philips, S. J., Wu, X., Chen, K., Shi, J., Shen, L., Xu, J., Feng, Y., O'Halloran, T. V., & Zhang, Y. (2021). CueR activates transcription through a DNA distortion mechanism. Nature chemical biology, 17(1), 57–64. https://doi.org/10.1038/s41589-020-00653-x